The University of Cambridge Naked Mole-Rat Initiative
The naked mole-rat (Heterocephalus glaber) is a mammal with a truly bizarre appearance, looking like an elongated cocktail sausage with large, protruding teeth.
Naked mole-rats live in large underground colonies of approximately 80 animals, which are dominated by a single breeding female, the queen; this social system is highly unusual in mammals, but is similar to that commonly observed in bees and termites and is termed eusocial1.
You can read more about the unusual biology of the naked mole-rat below and how members of the NMRI have contributed to our understanding of this fascinating species. If you wish to collaborate on a naked mole-rat project then in the first instance contact the Director of the NMR, Prof. Ewan St. John Smith.
In 2021, Ewan made a series of animations funded by the Dunhill Medical Trust to disseminate some of the magic of the naked mole-rat - you can see Episode 1 of Naked Mole Ravolt here: https://www.youtube.com/embed/kxSivZYJk5k.
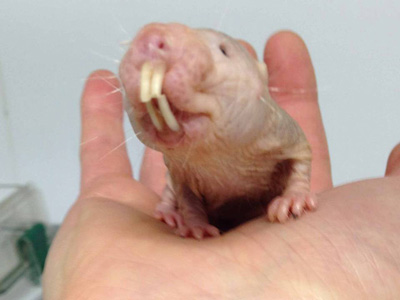
Over the last decade further physiological peculiarities of naked mole-rat physiology have come to light:
Extreme Longevity – naked mole-rats live until 30 years of age, whereas the longevity of similarly sized mice is 2-3 years; moreover, naked mole-rats display sustained good health into old age2,3
Cancer Resistance – naked mole-rats rarely spontaneously develop cancer4, but the question is why?
Insensitivity To Acid As A Noxious Stimulus – naked mole-rats respond normally to mechanical and thermal stimuli, but fail to perceive acid as noxious5, at least when applied subcutaneously, some acid-sensitivity remains intact in visceral sensory neurones6
Hypoxia Resistance – naked mole-rat brain tissue can withstand sustained periods of hypoxia and even anoxia7,8
While such phenomena are of great interest there has been little work identifying their causes, although there is clearly much that could be learned from the naked mole-rat that could aid advances2.
In 2014, scientists from the University of Cambridge Department of Pharmacology established the University of Cambridge Naked Mole-Rat Initiative (NMRI), which aims to bring together experts in different scientific areas with the overarching aim being to identify molecular explanations for the highly unusual physiology of this species. The majority of the work carried out by this initiative will be on established naked mole-rat cell lines.
An example of previous success in this area comes from a study by Dr Ewan St. John Smith, a founding member of the NMRI, which identified the molecular basis of naked mole-rat acid-insensitivity. It was shown that a variant in the voltage-gated sodium channel NaV1.7 is more greatly inhibited by acid in naked mole-rat pain-sensing neurones (nociceptors) compared to mouse: acid anaesthetises, rather than stimulates, naked mole-rat nociceptors9.
This work thus demonstrates the power of comparative physiology: through using standard rodent models the role of voltage-gated sodium channels in the acid-pain pathway had not been demonstrated, but by taking advantage of the naked mole-rat’s natural adaptation to its environment we were able to identify an important function of NaV1.7.
Interestingly, recent work from the NMRI has shown that colonic sensory nerves from naked mole-rats retain some acid sensitivity, albeit less than mouse colonic sensory nerves, but that acid only sensitises naked mole-rat colonic sensory nerves6; these data demonstrate a difference between somatic and visceral acid perception, as well as likely signalling differences underpinning neuronal sensitisation across species. Follow up work demonstrated that the naked mole-rat gastrointestinal tract displays reduced permeability10, understanding the mechanisms underpinning this low permeability could provide insights in conditions, such as Crohn's disease.
More recently Ewan was part of a study that demonstrated that naked mole-rats are highly resistant to hypoxia and anoxia due to their cells being able to efficiently utilise fructose to power energy production during periods of low oxygen8. This work enhances understanding of how nerve cells can function in the absence of oxygen and might lead to work that uncovers novel treatments to prevent brain damage in stroke patients.
Ewan’s lab have also recently identified appropriate housekeeping genes for the expression studies using naked mole-rat tissue11, shown that naked mole-rat cortical neurones are resistant to acid-induced cell death12, and in a study led by Prof. Matthew Mason’s lab in the Department of Physiology, Development and Neuroscience, an in depth anatomical description of the naked mole-rat’s peripheral auditory system was made13, which sheds light on the restricted hearing of naked mole-rats.
An area of research that has received a lot of attention is that of the naked mole-rat's resistance to cancer. In a collaboration between the labs of Dr Walid Khaled and Ewan, it was shown that contrary to previous reports, naked mole-rat cells could be transformed by well characterised oncogenes, i.e. their cancer resistance is likely non-cell autonomous, perhaps arising from a different extracellular environment and/or an altered immune response to cancer cells14; this story received a lot of media attention as you can read here and here. A recent collaboration with researchers at the Sanger Institute also identified that the rate of somatic mutations in naked mole-rats (a mechanism underlying some cancers) was much slower in naked mole-rats than in similarly sized mice15, a story that you could read about on the BBC.
References:
1. Jarvis, J.U. Science 212, 571–3 (1981).
2. Schuhmacher, L.-N., Husson, Z. & Smith, E.S. Open Acc Anim Phys 137 (2015).
3. Buffenstein, R. J Comp Physiol B 178, 439–45 (2008).
4. Delaney, M.A., Nagy, L., Kinsel, M.J. & Treuting, P.M. Vet Pathol 50, 607–621 (2013).
5. Park, T.J. et al. PLoS Biol 6, e13 (2008).
6. Hockley, J.R.F., Taylor, T.S., Callejo, G., Husson, Z.M. and Smith, E.S. Mol. Pain 16, 1-11 (2020).
7. Larson, J. & Park, T.J. Neurorep 20, 1634–1637 (2009).
8. Park, T.J. et al. Science 356, 307–311 (2017).
9. Smith, E.S.J. et al. Science 334, 1557–1560 (2011).
10. Aguilera-Lizarraga, J. et al Am. J. Physiol., 327, G188-201 (2024).
11. Schuhmacher, L.-N. & Smith, E.S.J. Mol. Brain 9, 97 (2016).
12. Husson, Z.M.A. & Smith, E.S.J. Mol. Brain 11, 26 (2018).
13. Mason, M.J., Cornwall, H.L. & Smith, E.S.J. PLOS ONE 11, e0167079 (2016).
14. Hadi, F. Kulaberoglu, Y., Lazarus, K.A., Bach, L., Ugur, R., Beattie, P., Smith, E. S. and Khaled, W.T. Nature, 583, E1 (2020).
15. Cagan et al, Nature, 604, 517-524 (2022).
Public Engagement
Ewan and others in his group are regularly involved in public engagement and widening participation initiatives, and the naked mole-rat provides a great example of being able to talk about how important it is to study basic biology to help understand normal physiological processes and what goes wrong in disease.
As well as giving talks at the Cambridge Science Festival, Cambridge BRAINFest and many schools, Ewan has been interviewed about his work for many media outlets, including: BBC Look East, BBC Radio Cambridgeshire and That's Cam TV, as well as the Danish newspaper Weekendavisen and Swedish radio.
Here is a piece that Ewan wrote for The Conversation and an interview with That's Cam TV, and of course you can see all 3 Episodes of Naked Mole Ravolt on YouTube. Here are links to videos Ewan made with Understanding Animal Research regarding animal husbandry of naked mole-rats and their use in cancer research.
Below you can read about the members of the NMRI, their expertise and current research interests.
University of Cambridge NMRI Members
Prof Ewan St. John Smith (Pharmacology)
NMRI role: Director and PI
Research Interests: Hypoxia/hypercapnia insensitivity, nociception and cancer resistance
Expertise: Electrophysiology, molecular biology, cell culture, immunohistochemistry and behaviour
Podcasts: With Science, The Physiological Society and the Naked Scientist
Prof. Walid T. Khaled (Pharmacology)
NMRI role: PI
Research Interests: Cancer development and heterogeneity
Expertise: Cancer biology, genetically engineered cancer models and genetic screens
Prof. Laura Itzhaki (Pharmacology)
NMRI role: PI
Research Interests: Cancer resistance and protein homeostasis
Expertise: Protein folding and stability, cancer therapeutics
Prof. Matt Mason (PDN)
NMRI role: PI
Research Interests: Structure and function of the middle ear in subterranean mammals
Expertise: Anatomy, biology of hearing.
Dr Janet Kumita (Pharmacology)
NMRI role: PI
Research Interests: Amyloid-related disorders and protein homeostasis
Expertise: Protein folding/misfolding, amyloid diseases
Dr Gabriel Balmus (UK Dementia Research Institute/Clinical Neurosciences)
NMRI role: PI
Research Interests: Neurodegeneration, DNA damage response
Expertise: Mouse models, CRISPR-Cas9 screening,
NMRI External Collaborators
Nicolas Cenac (INSERM, Toulouse University)
NMRI role: PI
Research Interests: Neuroimmune interactions, pain
Expertise: Lipidomics, neuronal functions, behaviour
NMRI Funding
We are grateful for the following support:
The Leona M. and Harry B. Helmsley Charitable Trust: New insights into Crohn's disease from studying Naked Mole Rat (2025 -27)
Dunhill Medical Trust: Naked neurones: how does the naked mole-rat brain stay healthy during ageing? (2020 - 23)
EMBO: Molecular mechanisms of neuronal tolerance to hypoxia in a naturally resistant mammal: the naked mole-rat (Fellowship awarded to Dr Zoé Husson, 2016 - 2018)
Isaac Newton Trust: Molecular mechanisms of neuronal tolerance to hypoxia in a naturally resistant mammal: the naked mole-rat (2016 - 2018)
Cancer Research UK: Determining and Exploiting the Anti-Cancer Properties of Naked Mole-Rat Hyaluronan (2016 - 2021)
NMRI Alumni
Dr Urriola-Muñoz (Postdoc, Smith Lab, Pharmacology, Cambridge, now Research Coordinator, Cambridge Centre for Physical Biology)
Dr Fazil Hadi (PhD student, Khaled Lab, Pharmacology, Cambridge, now scientist at AstraZeneca)
Dr Yavuz Kulaberoglu (Postdoc, Smith Lab, Pharmacology, Cambridge, now postdoc at UCL)
Becky Rickman (MPhil Student, Smith Lab, Pharmacology, Cambridge)
Jiwon Yi (MPhil student, Smith Lab, Pharmacology, Cambridge, now PhD student WashU)
Dr Zoé Husson (Postdoc, Smith Lab, Pharmacology, Cambridge, now Maitre de conférences, Université de Montpellier)
Dr Laura-Nadine Schuhmacher (PhD student, Smith Lab, Pharmacology, postdocs at UCL and Cardiff, now patent attorney)
NMRI Publications
Shepard, A., Lester, D.K., Troutman, S., Hoxha, S., Khaled, W.T., Smith, E.S., Park, T.J., Buffenstein, R., Du, D., Teng, M., Dengler-Crish, C.M., Tsai, K.Y., Flores, E.R., Ventura, A., and Kissil, J.K. An autochthonous model of lung cancer identifies requirements for cellular transformation in the naked mole-rat. Cancer Discovery, in press doi 10.1158/2159-8290.CD-25-0526
Peacock, J., Smith, E.S., Park, T.J. and Mason, M.J.† (2025) Ear morphology in the Damaraland mole-rat, Fukomys damarensis (Rodentia: Bathyergidae). Mammalian Biology, in press, doi 10.1007/s00359-025-01752-7.
Qiu, L. and Smith, E.S.† Artemin sensitises mouse (Mus musculus) and naked mole-rat (Heterocephalus glaber) sensory neurons in vitro. J Comp Physiol A, in press, doi 10.1007/s00359-025-01752-7
Drugachenok, P., Urriola-Munoz, P., Qiu, L., Han, Z., Smith, E.S., and Rahman, T.† (2025) Functional characterization of the store-operated calcium entry pathway in naked mole-rat cells.Open Biology, 15, 250052
Aguilera-Lizarraga, J.†, Ritoux, A., Bulmer, D.C. and Smith, E.S.† (2024) Intestinal barrier function in the naked mole-rat: an emergent model for gastrointestinal insights. Am J Physiol, 327, G188-201.
Urriola-Muñoz, P., Pattison, L.A., and Smith, E.S.† (2023) Dysregulation of ADAM10 shedding activity as a mechanism of cancer resistance in the naked mole-rat. J Cell Physiol, 238, 761-75.
Goyens, J., Baeckens, S., Smith, E.S., Pozzi, J., and Mason, M.J. (2022) Parallel evolution of semicircular canal form and sensitivity in subterranean mammals. J Comp Physiol A, 208, 627-40.
Fatima, I., Chen, G., Botchkareva, N.V., Sharov, A.A., Thornton, D., Wilkinson, H.N., Hardman, M.J., Grutzkau, A., de Magalhaes, J.P., Seluanov, A., Smith, E.S., Gorbunova, V., Mardaryev, A.N.†, Faulkes, C.G.†, and Botchkarev, V.A. (2022) Skin aging in long-lived naked mole-rats is accompanied by increased expression of longevity-associated and tumor suppressor genes. J Inv Derm, 142, 2853-63.
Cagan, A., Baez-Ortega, A., Brzozowska, N., Abascal, F., Coorens, T.H., Sanders, M.A., Lawson, A.R., Harvey, L.M., Bhosle, S.G., Jones, D., Alcantara, R.E., Butler, T.M., Hooks, Y., Roberts, K., Anderson, E., Flach, E., Spiro, S., Januszczak, I., Wrigglesworth, E., Perkins, M.W., Deaville, R., Druce, M., Bogeska, R., Milsom, M.D., Neumann, B., Gorman, F., Constantino-Casas, F., Peachey, L., Bochynska, D., Smith, E.S., Gerstung, M., Campbell, P.J., Murchison, E.P., Stratton, M.R., and Martincorena, I. (2022) Somatic mutation rates scale with lifespan across mammals. Nature, 604, 517-24
Smith, E.S., Park, T.J., Holmes, M.M. and Buffenstein, R.† (2021). Some Exciting Future Directions for Work on Naked Mole-Rats. Adv Exp Med Biol, 1319, 409-420.
Hadi, F., Smith, E.S.†, and Khaled, W.T.† (2021). Naked Mole-Rats: Resistant to Developing Cancer or Good at Avoiding It? Adv Exp Med Biol, 1319, 341-352.
Park, T.J.†, Smith, E.S.., Reznick, J., Bennett, N.C., Applegate, D.T., Larson, J., Lewin, G.R. (2021). African Naked Mole-Rats Demonstrate Extreme Tolerance to Hypoxia and Hypercapnia. Adv Exp Med Biol, 1319, 255-269.
Lewin, G.R.†, Smith, E.S., Reznick, J., Debus, K., Barker, A.J. and Park, T.J. (2021). The Somatosensory World of the African Naked Mole-Rat. Adv Exp Med Biol, 1319, 197-220.
Vice, E.N., Lagestee, S., Browe, B.M., Deb, D., Smith, E.S. and Park, T.J.† (2021). Sensory Systems of the African Naked Mole-Rat. Adv Exp Med Biol, 1319, 137-156.
Hadi, F., Kulaberoglu, Y., Lazarus, K., Beattie, P., Smith, E.S. and Khaled, W.T. Transformation of naked mole-rat cells. Nature, 583, E1-E7
Smith, E.S. and Park, T.J. (2020). Neurobiology: Crowdsourcing CO2 to conserve brain energy. Curr Biol, 30, R649-651
Hacker, L., Brunker, J., Smith, E.S., Gozales, I.Q. and Bohndiek, S.E. (2020). Photoacoustics resolves species-specific differences in haemoglobin concentration and oxygenation. J Biomed Opt,
Browe, B.M., Olsen, A.R., Ramirez, C., Rickman, R.H., Smith, E.S. and Park, T.J. (2020). The naked mole-rat has a functional purinergic pain pathway despite having a non-functional peptidergic pain pathway. Neurobiol Pain, 8, 100047
Smith, E.S., Park, T.J. and Lewin, G.R. Independent evolution of pain insensitivity in African mole-rats: origins and mechanisms. J Comp Physiol A, 206, 313-325
Hockley, J.R.F., Taylor, T.S., Callejo, G., Husson, Z.M. and Smith, E.S. Acid and inflammatory sensitisation of naked mole-rat colonic afferent nerves. Mol. Pain 16, 1-11 (2020)
Kulaberoglu, Y., Bhushan, B., Hadi, F., Chakrabarti, S., Khaled, W.T., Rankin, K., Smith, E.S. and Frankel, D. (2019) The material properties of naked mole-rat hyaluronan. Sci Reports, 9, 6632
Husson, Z. and Smith, E. S (2018) Naked mole-rat cortical neurons are resistant to acid-induced cell death. Mol Brain, 11, 26
Schuhmacher, L. N., Callejo, G., Srivats, S. and Smith, E. S. (2018). Naked mole-rat acid-sensing ion channel 3 forms nonfunctional homomers, but functional heteromers. J Biol Chem, 293, 1756-1766.
Park, T. J., Reznick, J., Peterson, B. L., Blass, G., Omerbasic, D., Bennett, N.C. Kuich, P.H.J.L, Zasada, C., Browe, B.M., Hamann, W., Applegate, D.T., Radke, M.H., Kosten, T., Lutermann, H., Gavaghan, V., Eigenbrod, O., Bégay, V., Amoroso, V.G., Govind, V., Minshall, R.D., Smith, E.S., Larson, J., Gotthardt, M., Kempa, S. and Lewin, G.R. (2017) Fructose-driven glycolysis supports anoxia resistance in the naked mole-rat. Science, 356, 307-311.
Schuhmacher, L. N. and Smith, E. S. (2016). Expression of acid-sensing ion channels and selection of reference genes in mouse and naked mole rat. Mol. Brain, 9, 97.
Mason, M. J., Cornwall, H. L. and Smith, E. S. (2016). Ear Structures of the Naked Mole-Rat, Heterocephalus glaber, and Its Relatives (Rodentia: Bathyergidae). PLoS One, 11, e0167079.
Omerbasic, D., Smith, E. S., Moroni, M, Eigenbrod, O., Homfeld, J., Reznick, J., Bennett, N. C. Faulkes, C., Selbach, M, and Lewin, G. R. (2016). Hypofunctional TrkA accounts for the absence of pain sensitization in the African naked mole-rat. Cell Rep., 17, 748-758
Schuhmacher, L. N., Husson, Z. and Smith, E. S. (2015). The naked mole-rat as an animal model in biomedical research: current perspectives. Open Access Animal Physiology, 7, 137-48.
