Biography
Dr Zinzalla obtained her first class honours Laurea in Chemistry (equivalent to MChem) from the University of Milan (Italy) and carried out her PhD studies with Prof. Stefano Maiorana and Dr Clara Baldoli also at the University of Milan. Her research focused on developing new metal complex conjugates of Peptide Nucleic Acids (PNAs) as biosensors, and she investigated novel trace-less linkers for solid-phase synthesis of drug-like small molecules in collaboration with GlaxoSmithKline.
In 2004 she was awarded a Marie Curie EIF Individual Research Fellowship and appointed as postdoctoral research assistant at the Department of Chemistry of the University of Cambridge (UK) with Prof. Steven Ley. Within the Ley group she initiated a project focused on exploring the role of molecular diversity in drug discovery, with the design of natural product-like compounds as novel therapeutic agents.
In June 2006 she joined the School of Pharmacy, University College London (UK) as a senior research fellow funded by Cancer Research UK (CR UK), within the CR UK Drug Discovery unit headed by Prof. David E. Thurston. As a co-principal investigator of a CR UK Small Molecule Drug Discovery Initiative program, she led the research aimed at discovering protein-protein interaction inhibitors of transcriptional factors such as Hypoxia Inducible Factor 1 (HIF-1) and Signal Transducer and Activator of Transcription 3 (STAT3 ).
In January 2012 she took up the position at the Karolinska Institutet and started her own research group as an assistant professor in chemical biology and principal investigator at the MTC Department and within the Centre for Advanced Cancer Therapies (ACT). In summer 2024 she joined the Department of Pharmaceutical Biosciences, Uppsala Biomedical Centre, Uppsala University.
At the University of Cambridge Dr Giovanna Zinzalla is a long-term visiting academic/PI in the Department of Pharmacology, where she carries out some of her research and lectures for the third-year/Part II Pharmacology course.
She is also a Bye-Fellow in Pharmacology, and Director of Studies in Natural Sciences (Biological) of the Lucy Cavendish College, where she is responsible for the students' academic progress and teaches pharmacology to students taking Natural Sciences Tripos and Medicine courses. At the College she is also an undergraduate Tutor providing pastoral support for the students' welfare.
Research
HOW TRANSCRIPTIONAL REGULATORS CONTROL GENE EXPRESSION PROGRAMS and CELL FATE in NORMAL and MALIGNANT CELLS
How the transcriptional machinery orchestrates gene expression programs during development, renewal and in response to environmental signals is one of the outstanding questions in biology. I tackle this fundamental problem by focusing on determining at the molecular level the mechanisms regulating the interactions between transcription factors and their interactions with co-factors, especially with epigenetic regulators.
My research approaches encompass the use of chemistry, biochemistry, biophysics and structural biology. The ultimate goal is to develop therapeutic agents to treat human diseases, and new approaches for regenerative medicine.
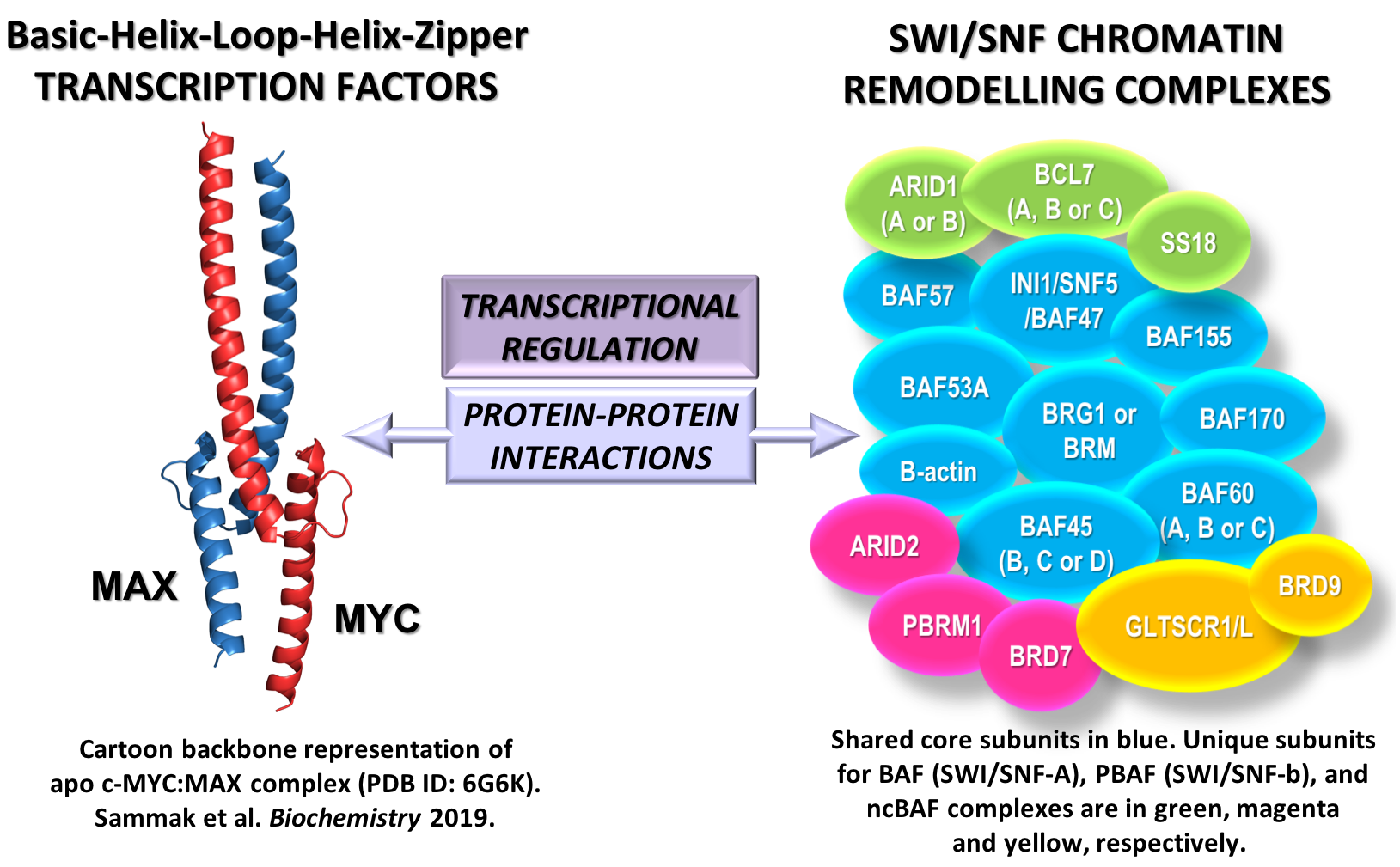
We have been focusing on two key molecular machineries: (1) MYC and other members of its Basic-Helix-Loop-Helix-Zipper (bHLHZip) network of transcription factors; and (2) the SWI/SNF (BAF) chromatin remodelling complexes.
We work on determining: how the MYC transcription factor cooperation leads to gene repression; and how the MYC extended network of bHLHLZ transcription factors, i.e., the c-MYC like MONDOA/ChREBP paralogs, regulate metabolism.
Our projects on the SWI/SNF complexes are currently directed to understand: how these chromatin remodelling complexes control MYC gene expression; and how the BAF60 and the BAF155/170 subunits, respectively, mediates interactions with diverse transcription factors with roles in cellular processes, such as regeneration, cardiac development and metabolism. We are also exploring ways to target the SWI/SNF complexes for the development of new therapies.
RESEARCH COLLABORATIONS (alphabetical order): Prof. Don Ayer, Huntsman Cancer Institute, University of Utah, USA; Dr Mark Bycroft, University of Cambridge, Cambridge, UK; Prof. Gerard Evan, Crick Institute, UK; Prof. Laura Itzhaki, University of Cambridge, Cambridge, UK; Prof. Lars-Gunnar Larsson, Uppsala University, Sweden; Prof. Mark Rubin MD, Bern Center for Precision Medicine (BCPM), University of Bern, Switzerland; Prof. Sophia Yaliraki, Imperial College London, UK.
Figure 1. The complex interplay between transcription factors and chromatin remodelling machines: on the left, the crystal structure of the bHLHZip C-terminal domain of human protein c-MYC (in red) bound to its obligatory partner MAX (in blue), and on the right, a schematic representation of the subunits of SWI/SNF complexes. Shared core subunits of the SWI/SNF complexes are in blue. Unique subunits belonging to BAF (SWI/SNF-A), PBAF (SWI/SNF-b), and ncBAF (non-canonical SWI/SNF) complexes are in green, magenta and yellow, respectively. The interplay is mediated by protein-protein interactions and mutual transcriptional regulation to control each other levels of protein expression. Illustration: Giovanna Zinzalla
Publications
Biophysical and Structural Methods to Study the bHLHZip Region of Human c-MYC Zinzalla G Methods in molecular biology (Clifton, N.J.) 2021;2318():21-43
SWI/SNF subunit BAF155 N-terminus structure informs the impact of cancer-associated mutations and reveals a potential drug binding site Allen MD, Freund SMV, Bycroft M, Zinzalla G Communications biology 2021;4(1):528-
Structure of the BRK domain of the SWI/SNF chromatin remodeling complex subunit BRG1 reveals a potential role in protein-protein interactions. Allen MD, Bycroft M, Zinzalla G Protein Sci. 2020 Apr;29(4):1047-1053.
Crystal Structures and Nuclear Magnetic Resonance Studies of the Apo Form of the c-MYC:MAX bHLHZip Complex Reveal a Helical Basic Region in the Absence of DNA. Sammak S, Hamdani N, Gorrec F, Allen MD, Freund SMV, Bycroft M, et al Biochemistry 2019 Jul 23;58(29):3144-3154.
The structure of INI1/hSNF5 RPT1 and its interactions with the c-MYC:MAX heterodimer provide insights into the interplay between MYC and the SWI/SNF chromatin remodeling complex. Sammak S, Allen MD, Hamdani N, Bycroft M, Zinzalla G FEBS J. 2018 11;285(22):4165-4180
A selective high affinity MYC-binding compound inhibits MYC:MAX interaction and MYC-dependent tumor cell proliferation. Castell A, Yan Q, Fawkner K, Hydbring P, Zhang F, Verschut V, et al Sci Rep 2018 Jul;8(1):10064
The SWI/SNF Subunit INI1 Contains an N-Terminal Winged Helix DNA Binding Domain that Is a Target for Mutations in Schwannomatosis. Allen MD, Freund SM, Zinzalla G, Bycroft M Structure 2015 Jul;23(7):1344-9
Tetracycline analogues with a selective inhibitory effect on HIF-1alpha Bendiabdellah Y, Rahman, KM, Uranchimeg B, Nahar KS, Antonow D, Shoemaker R.H., Melillo G, Zinzalla G, Thurston, DE Med. Chem. Commun., 2014,5, 923-926.
Observation of unphosphorylated STAT3 core protein binding to target dsDNA by PEMSA and X-ray crystallography. Nkansah E, Shah R, Collie GW, Parkinson GN, Palmer J, Rahman KM, et al FEBS Lett. 2013 Apr;587(7):833-9
Small-molecule inhibition of c-MYC:MAX leucine zipper formation is revealed by ion mobility mass spectrometry. Harvey SR, Porrini M, Stachl C, MacMillan D, Zinzalla G, Barran PE J. Am. Chem. Soc. 2012 Nov;134(47):19384-92
Facile nucleophilic substitution at the C3a tertiary carbon of the 3a-bromohexahydropyrrolo[2,3-b]indole scaffold. Villanueva-Margalef I, Thurston DE, Zinzalla G Org. Biomol. Chem. 2010 Dec;8(23):5294-303
Facile oxidation of electron-poor benzo[b]thiophenes to the corresponding sulfones with an aqueous solution of H2O2 and P2O5. Antonow D, Marrafa T, Dawood I, Ahmed T, Haque MR, Thurston DE, et al Chem. Commun. (Camb.) 2010 Apr;46(13):2289-91
Natural-product-like spiroketals and fused bicyclic acetals as potential therapeutic agents for B-cell chronic lymphocytic leukaemia. Milroy LG, Zinzalla G, Loiseau F, Qian Z, Prencipe G, Pepper C, et al ChemMedChem 2008 Dec;3(12):1922-35