Structure and function of biological macromolecules examined using the atomic force microscope
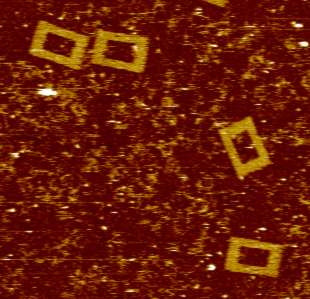
 Since 1996 we have been using atomic force microscopy (AFM) as a tool to study various aspects of cell and molecular biology (work being mainly with Professor Mike Edwardson). Our aim has always been to use AFM as a tool, normally to complement more mainstream molecular biological techniques, to allow direct observation of macromolecular features that can only be studied indirectly using these mainstream methods. We have worked on a number of different systems, including elucidation of mechanisms of exocytosis; membrane structure and function and molecular interactions with membranes; structural effects of drugs that intercalate into DNA; and the oligomeric assembly of receptors and ion channels and have published numerous papers describing work using AFM to study molecular biological topics since 1997. For some years, our work was predominately focused on these studies of the architecture of therapeutically significant receptors and ion channels and also on investigations of the mechanisms of action of DNA restriction-modification enzymes. However, in recent years we have expanded our interests and have also published on protein interactions with lipid bilayers, and the structure of G-quadruplex DNA (which adopts unusual conformations - see figure). We also have long-standing interest in high-speed fast-scanning AFM.
Since 1996 we have been using atomic force microscopy (AFM) as a tool to study various aspects of cell and molecular biology (work being mainly with Professor Mike Edwardson). Our aim has always been to use AFM as a tool, normally to complement more mainstream molecular biological techniques, to allow direct observation of macromolecular features that can only be studied indirectly using these mainstream methods. We have worked on a number of different systems, including elucidation of mechanisms of exocytosis; membrane structure and function and molecular interactions with membranes; structural effects of drugs that intercalate into DNA; and the oligomeric assembly of receptors and ion channels and have published numerous papers describing work using AFM to study molecular biological topics since 1997. For some years, our work was predominately focused on these studies of the architecture of therapeutically significant receptors and ion channels and also on investigations of the mechanisms of action of DNA restriction-modification enzymes. However, in recent years we have expanded our interests and have also published on protein interactions with lipid bilayers, and the structure of G-quadruplex DNA (which adopts unusual conformations - see figure). We also have long-standing interest in high-speed fast-scanning AFM.
We have three Bruker atomic force microscopes in the laboratory, including a FastScan instrument, recently acquired, which allows very rapid capture of images, allowing near-real time imaging of molecular activity. We are happy to make this instrument available to the wider research community.
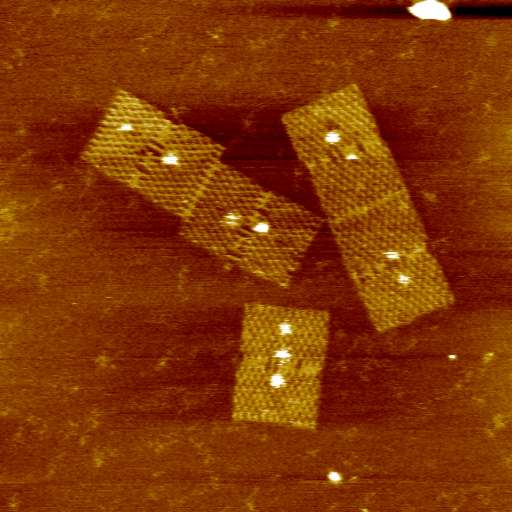
At present we are looking principally at two systems, each using DNA origami (which we are studying in collaboration with Professor Hirsoshi Sugiyama and Dr Masayuki Endo at Kyoto University and Dr David Dryden at the University of Edinburgh):
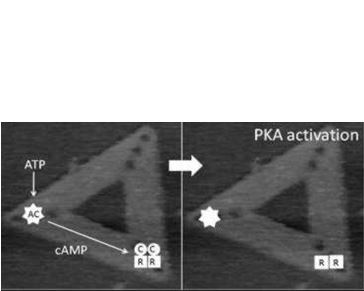
First, the spatial relationship between the differing elements of the adenylyl cyclase (AC)/protein kinase A (PKA)/phosphodiesterase (PDE) signalling system. The successful operation of this system requires correct positional arrangement of the different components to allow, first, cAMP to be generated, then to allow it to interact with PKA, and finally, PDE must be in appropriate proximity to allow regulated degradation of cAMP. The exact geometrical relation between these components still is unknown. We are generating origami scaffolds, and attach each of the elements of the AC/PKA/PDE system in differing orientations and examine the role of orientation on activation by looking at changes in the structure of the PKA, which alters during the signalling process. The real-time imaging allowed by fast-scan AFM provides a powerful new technique to study the process directly.
 |
 |
 |
|---|
The second set of experiments also addresses a question that has proved difficult to resolve using other methods. We are making DNA origami scaffolds that bear sequences of DNA that bear recognition sites for DNA-binding proteins (for example proteins involved in gene transcription and DNA replication and repair). We are using these to quantify the dynamics and kinetics of protein interaction and target location on the DNA. How do proteins rapidly and accurately find their relatively short target sequences on an enormously long DNA molecule? This topic has exercised interest for many years, but the data so far available remains inconclusive and as with the other strand of our work real-time imaging allowed by fast-scan AFM makes direct observation of dynamic events possible.