Our research seeks to understand more about the cell cycle, and how cell division and cell fate decisions are shaped by ubiquitin-mediated signalling. Much of our focus is on the major cell cycle regulator Aurora A kinase (AURKA), a target of ubiquitin-dependent proteolysis by the APC/C-FZR1 ubiquitin ligase. Overexpression of AURKA is strongly associated with cancer progression. Alongside projects to understand the importance of AURKA regulation to cell cycle control and cancer, we are investigating new methods to manipulate and measure AURKA stability inside cells. We are also developing tools that will become essential for the therapeutic exploitation of intracellular protein stability. With the rapid growth in targeted protein degradation as a new therapeutic modality, this is a new and still unexplored frontier in pharmacology.
Please see our Publications page for details of our recent papers. Below, some of our lab members have used their time away from the bench during the Covid-19 crisis to generate graphical abstracts of their projects.
Cynthia Okoye: Toward the design of APC/C E3 ligase binding peptides and PROTACs (collaboration with Itzhaki lab).
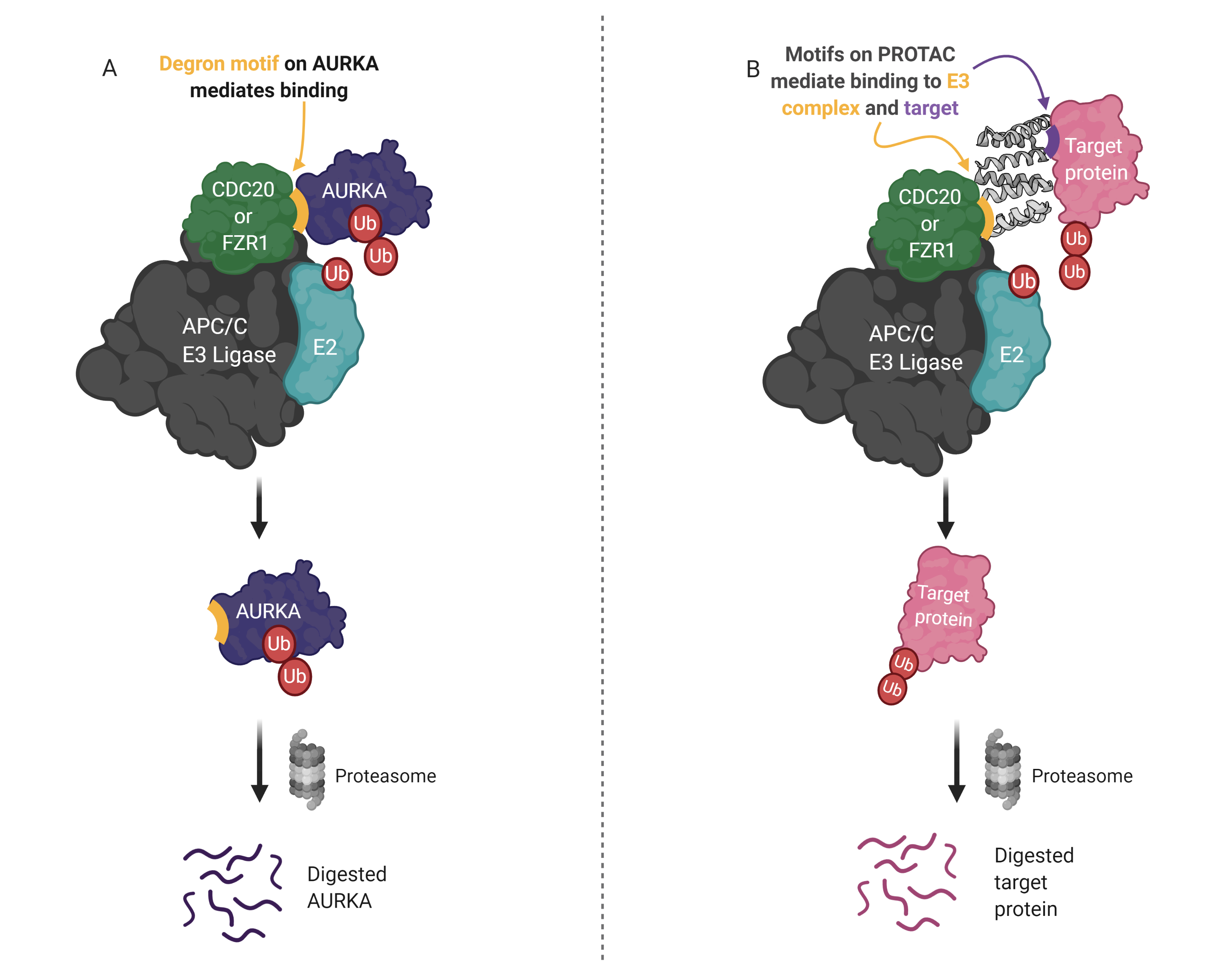
PROTACs are attractive therapeutics because they employ an event-driven pharmacology to reduce the cellular levels of proteins that are implicated in disease. Current PROTACs mostly utilize VHL, CRBN, MDM2 or cIAP1 E3 ligases, meanwhile, there are more than 600 E3s in humans. This project aims to design PROTACs that harness the anaphase-promoting complex (APC/C), which is a key player in the regulation of mitotic proteins including PTTG1, NEK2A, AURKA and many more. Natural substrates of the APC/C often contain short linear motifs (SLiMs), termed degrons, that serve as recognition sequences for binding and ubiquitination. Consequently, the design of APC/C-binding peptides in this project utilizes degrons found in AURKA and other well-characterized APC/C substrates. The designed peptides are tested in cell-based degradation assays using time-lapse quantitative fluorescence imaging. Novel degron combinations that yield significant degradation during mitotic exit in cells have been identified and will be used to build protein PROTACs that harness the APC/C.
Camilla Ascanelli: Fluorescent protein timers to characterise Myc stability (collaboration with AstraZeneca)
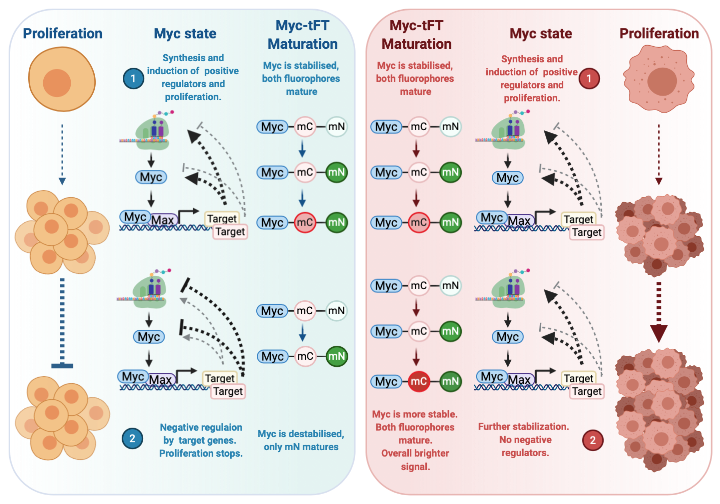
Myc is a transcription factor with oncogenic activity. In healthy cells, the MYC gene is switched on as a consequence of pro-growth signals. The resulting protein heterodimerizes with its DNA binding partner MAX to promote transcription of a number of genes involved in proliferation. The products of some of these genes positively feedback to increase Myc expression and protein stability. When the required proliferation has been satisfied, excess Myc leads to induction of proteins that negatively regulate its own expression and stability allowing cells to halt Myc-induced proliferation. This balance in positive and negative feedback loops is essential for controlled cellular division and often lost in cancer, where the positive feedback is amplified, and negative feedback suppressed. To investigate this balance in Myc regulation, we take advantage of fluorescent protein timers that consist of two fluorophores with differing maturation times fused to Myc: mCherry (mC) and mNeonGreen (mN). An unstable Myc protein has a short half-life and only allows maturation of mN. A stable Myc protein, which has a longer half-life, allows maturation of both fluorophores that emit light. Further stabilisation, such as that seen in a cancer setting, will produce even brighter signals from both mature fluorophores.
Roberta Cacioppo:
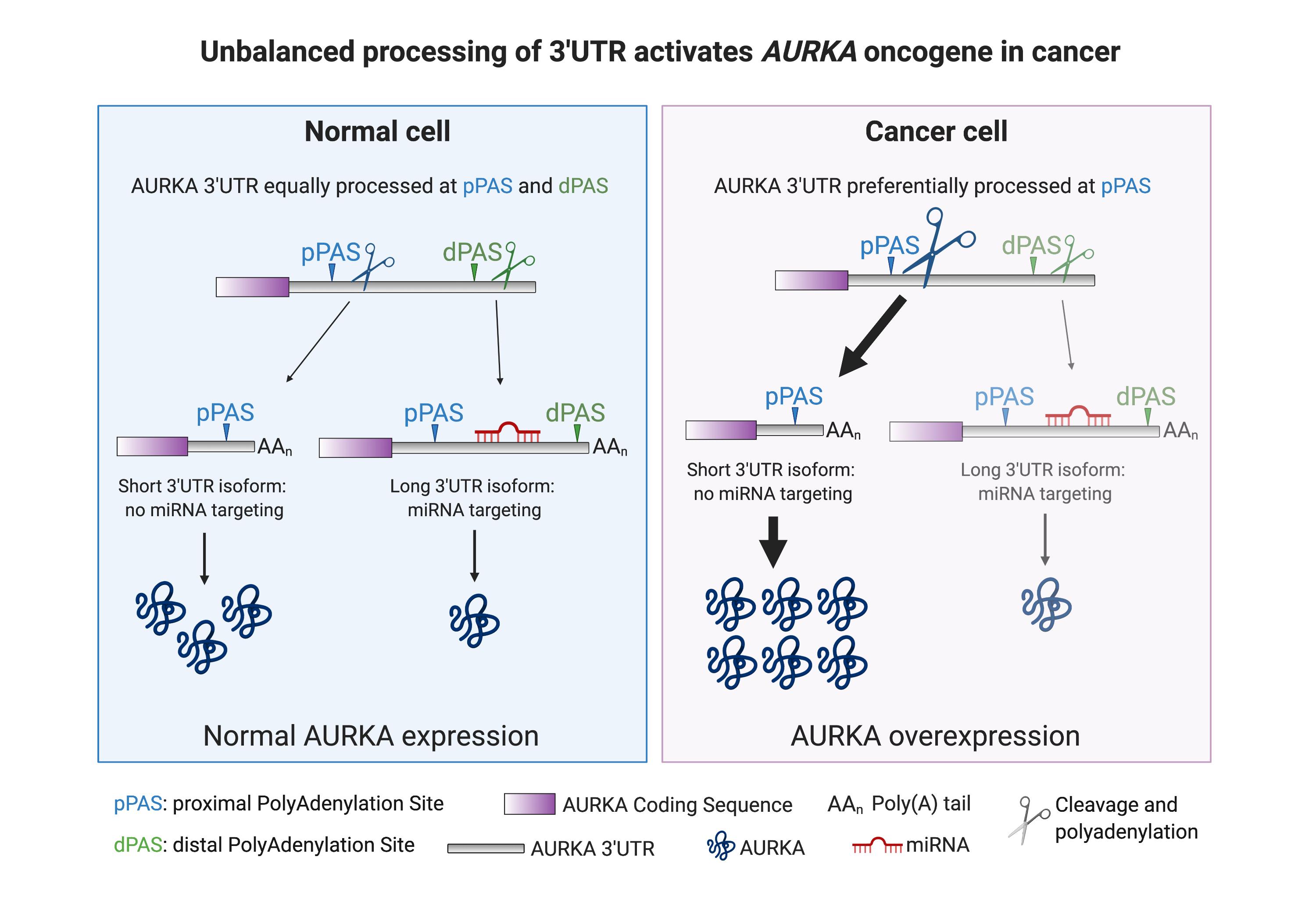
Aurora kinase A (AURKA) is a well-known regulator of the cell cycle as it promotes mitosis entry and progression. Its role as oncogene is nowadays widely established. The 3’Untranslated Region (UTR) of AURKA mRNA contains a proximal (p) and a distal (d) PolyAdenylation Site (PAS), which can be alternatively cleaved and polyadenylated resulting in a short or long 3’UTR isoform, respectively. Protein expression from the long isoform is potentially limited by miRNA targeting. Normally, AURKA 3’UTR is equally processed at both PASs, resulting in balanced AURKA protein production. In cancer cells, AURKA 3’UTR is preferentially processed at pPAS, causing AURKA protein overexpression, which underlies activation of AURKA oncogene.
