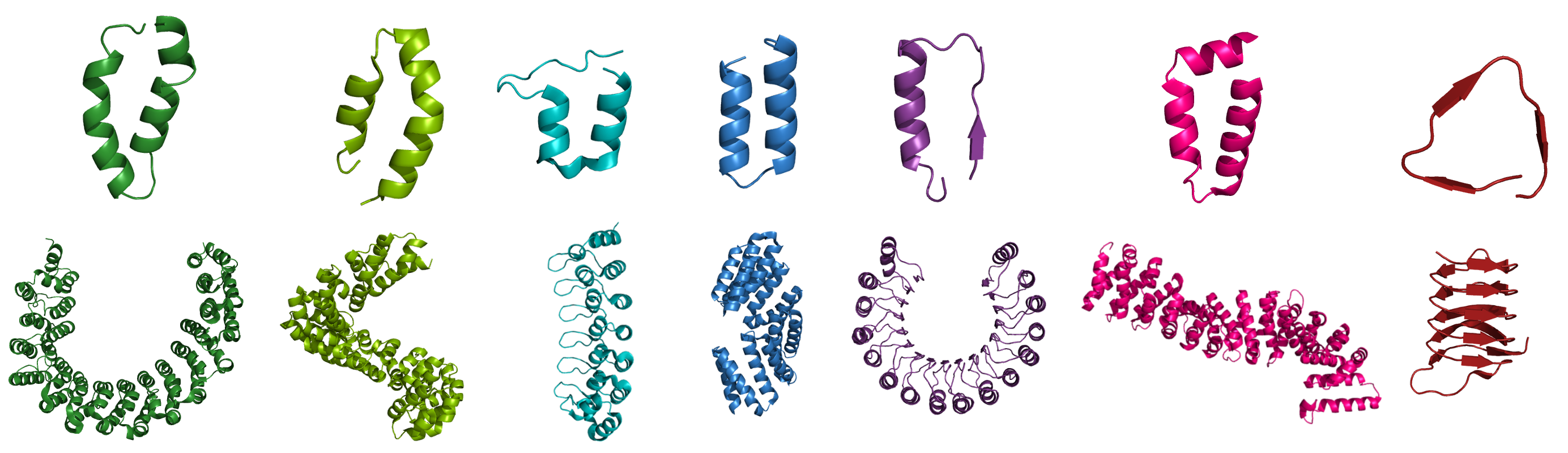
A major focus of our research is a class of proteins known as tandem-repeat proteins. They comprise small structural units repeated multiple times in tandem to form non-globular, elongated and superhelical structures that present extended scaffolds for molecular recognition. There is a large variety of repeat types ranging from purely helical or beta-sheet to a mix of both. Consequently, they give rise to many different geometrical shapes, which in turn confer different biochemical and biophysical properties and functions. thereby presenting an extended scaffold for molecular recognition. The term ‘scaffold’ implies a rigid architecture; however, as suggested by their spring-like shapes, it is thought that repeat arrays utilise much more dynamic and elastic modes of action. For example: stretching and contraction motions to regulate the activity of a bound enzyme; reversible nanosprings to operate ion channels; transporters that wrap around their cargoes to carry them in and out of the nucleus.
We use a battery of techniques spanning protein engineering, single-molecule and biophysics methods, biochemistry, and analysis in cellulo and in silico. These techniques, in combination with site-directed mutagenesis allow us to probe mechanisms and explore the fundamental laws that govern them. At the same time, we are also investigating novel approaches to therapeutics, motivated by what we know about the underlying principles of repeat-protein structure and function.
Structure, function and mechanics of repeat proteins
One major aim of our current research is to define the conformational transitions that repeat proteins undergo during the different stages of their life cycle - biosynthesis, folding, localisation, assembly and degradation - and how they are guided by the cellular machinery. Due to their modular architecture and spring-like shapes, it is proposed that repeat arrays utilise much more dynamic and elastic modes of action than globular proteins. For example: stretching and contraction motions to regulate the activity of a bound enzyme; acting as reversible nanosprings to operate ion channels; proteins that wrap around their cargoes to transport them between the nucleus and cytoplasm.
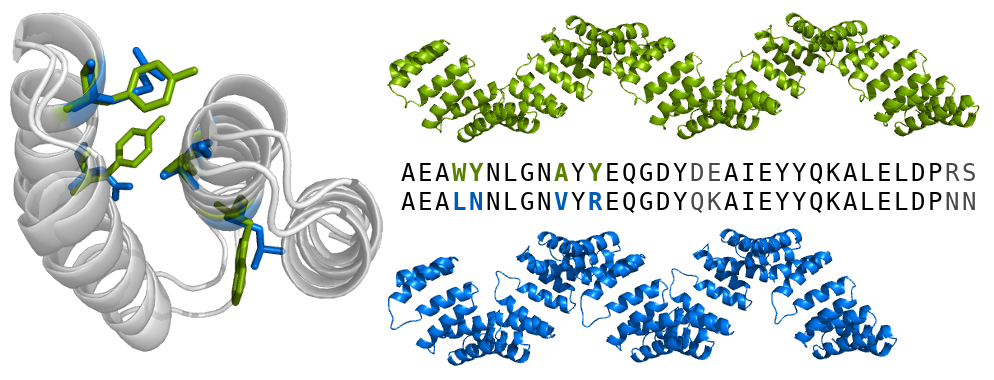
Our group and others have shown that the simple modular, 1D-like architecture of repeat proteins gives them distinctive properties compared with globular proteins and makes it uniquely straightforward to map the energetics of their structures and rationally redesign their stability, folding and molecular recognition.
This class of proteins is thus an exceptionally sensitive and versatile tool that we are now in a position to exploit to dissect otherwise intractable cellular mechanisms.
Banerjee A, Mathew S, Naqvi MM, Yilmaz SZ, Zacharopoulou M, Doruker P, Kumita JR, Yang SH, Gur M, Itzhaki LS, Gordon R, Bahar I. Influence of point mutations on PR65 conformational adaptability: Insights from molecular simulations and nanoaperture optical tweezers. Sci Adv. 2024 May 31;10(22):eadn2208. doi: 10.1126/sciadv.adn2208.
Ventura C, Banerjee A, Zacharopoulou M, Itzhaki LS, Bahar I. Tandem-repeat proteins conformational mechanics are optimized to facilitate functional interactions and complexations. Curr Opin Struct Biol. 2024 Feb;84:102744. doi: 10.1016/j.sbi.2023.102744.
Ripka JF, A Perez-Riba, PK Chaturbedy, LS. Itzhaki. Testing the length limit of loop grafting in a helical repeat protein. Curr. Res. Struct. Biol., 3(1): 30-40, 2021.
Perez-Riba A, Synakewicz M, Itzhaki LS. Opinion piece: Folding cooperativity and allosteric function in the tandem-repeat protein class. Phil. Trans. R. Soc. B, 373:20170188, 2018.
Molecular therapeutics, targeted protein degradation, and biomolecular condensates
We are focusing on repeat proteins that play important roles in disease, in particular cancer, and we are using the insights we obtain to develop therapeutic strategies for targeting them. In our group, we are exploring three different approaches to target our proteins of interest: (i) chemically constrained peptides, (ii) designed repeat proteins, and (iii) nanobodies (single-domain antibodies). Targets include the substrate-binding subunits of oncogenic E3 ubiquitin ligases and the substrate-recognition domain of the poly(ADP) ribose polymerase tankyrase. In other projects, we are engineering new functions into ultra-stable consensus-designed repeat proteins with the aim to disrupt aberrant protein-protein interactions. To remove proteins upregulated in disease, we are developing degraders such as bioPROTACs (chimeric E3 ubiquitin ligases) and exploiting short linear motifs to direct targets to proteasome and lysosome pathways for degradation. Lastly, we are exploring the use of biomolecular condensates and nanoparticles to concentrate our molecular therapeutics with their targets and thereby enhance potency.
Eapen R, Okoye C, Stubbs C, Schimpl M, Tischer T, McCall E, Zacharopoulou M, Ferrer F, Barford D, Spring D, Lindon C, Phillips C, Itzhaki LS. Development of D-box peptides to inhibit the Anaphase Promoting Complex/Cyclosome. bioRxiv preprint doi:https://doi.org/10.1101/2024.04.30.590460.
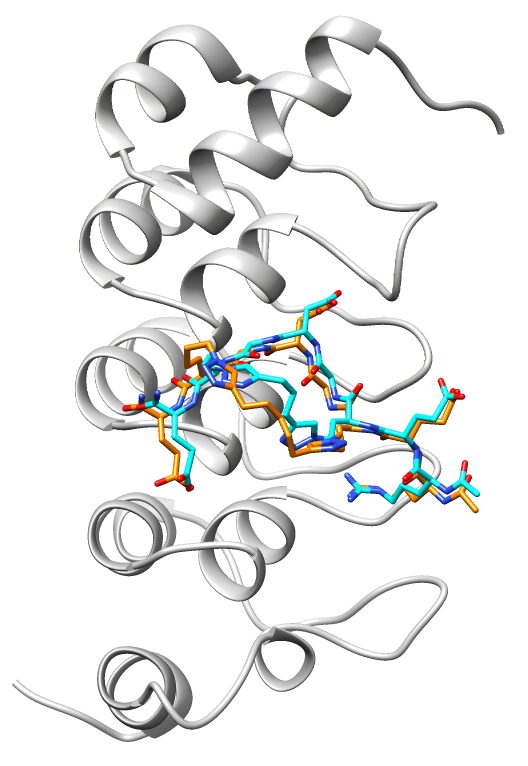
King HR, Bycroft M, Nguyen TB, Kelly G, Vinogradove AA, Rowling PJE, Stottf K, Ascher DB, Suga H, Itzhaki LS, Artavanis-Tsakonasa K. Targeting the Plasmodium falciparum UCHL3 ubiquitin hydrolase using chemically constrained peptides. PNAS 2024 Vol. 121 No. 21 e2322923121 https://doi.org/10.1073/pnas.2322923121.
Ng TLC, Hoare MP, Maristany MJ, Wilde EJ, Sneideris T, Huertas J, Agbetiameh BK, Furukawa M, Josep JA, Knowles TPJ, Collepardo-Guevara R, Itzhaki LS, Kumita JR. Tandem-repeat proteins introduce tuneable properties to engineered biomolecular condensates. bioRxiv preprint doi: https://doi.org/10.1101/2024.04.16.589709.
Burbidge O, MW Pastok, SL Hodder, G Zenkevičiūtė, MEM Noble, JA Endicott, LS Itzhaki. Nanobodies restore stability to cancer-associated mutants of tumor suppressor protein p16INK4a.
Zenkevičiūtė G, Xu W, Iegre J, Seki H, Tan YS, Rowling PJE, Ferrer F, Verma C, Spring DR, Laman H, Itzhaki LS. Development of constrained peptide inhibitors targeting an oncogenic E3 ubiquitin ligase. bioRxiv preprint doi:https://doi.org/10.1101/2023.04.27.535981.
Diamante A, P Chaturbedy, P Rowling, J Kumita, RS Eapen, SH McLaughlin, M de la Roche, A Perez-Riba, LS Itzhaki. Engineering mono- and multi-valent inhibitors on a modular scaffold. Chem. Sci., 12(3):880-895, 2021.
Xu W, YH Lau, G Fischer, YS Tan, A Chattopadhyay, M de la Roche, M Hyvönen, CS Verma, DR Spring, LS Itzhaki. Macrocyclized extended peptide Inhibiting the substrate-recognition domain of tankyrase. J. Am. Chem. Soc., 139(1):2245-2256, 2017.
Designed repeat proteins as tension sensors for interrogating mechanical forces in the cell
Forces are key to a wide variety of cellular processes, and It is clear that specific proteins must be able to sense mechanical signals and convert them into biological responses. However, a molecular-level understanding of such processes is very limited, as accurate measurement of the forces poses significant challenges, in particular because it is currently not possible for us to build force sensor with tailored mechanical characteristics. We believe that repeat proteins should be able to address this challenge. Our group and others have shown that these proteins behave like molecular “nanosprings” and, because of the modular structure, they can put together like Lego from small structural motifs into large arrays. Using these nanosprings, we will produce a toolkit of force sensors that can be customised for diverse in cellulo applications.
Synakewicz M, RS Eapen, A Perez-Riba, D Bauer, A Weißl, G Fischer, M Hyvönen, M Rief, LS Itzhaki, J Stigler. Consensus tetratricopeptide repeat proteins are complex superhelical nanosprings.